Coursework Summer 2014/2015
Varování:
This is not the current assignment.
Proteins
The topic of this year’s semester work are proteins. Your task is to develop OpenSCAD module, which generates protein models, which match given parameters, other part of your assignment is to correctly prepare the final model which will come out of your modul to print, last not least is to print it and perform all necessary actions afterwards.
Protein generator
Develop OpenSCAD modul protein which keeps given modul interface, which will render model of protein depending on given parameters.
module protein(aminos=[],bindings=[]) { ... }Vector (array) aminos contains definition of amino-acids. One amino-acid is represented by one ball with given center location and radius. Data are stored in four-element vectors [x,y,z,r], where first 3 elements describes location of ball center and the last element is radius. Vector aminos is vector of vectors:
aminos = [[x1, y1, z1, r1], [x2, y2, z2, r2], ...];
Vector bindings contains two-elements vectors (pair) of index of vector aminos. These pairs of amino-acids shall be connected by linkage, which will be described down bellow.
bindings = [[0, 1], [1, 8], ...]; // example
Linkage of two animo-acids is created by using center ball, placed by it’s center exactly to the center of two centers of two amino-acids, with radius of avg(ra,rb)/2, where avg is arithmetic average. ra and rb are radiuses of two amino-acids, which you are trying to link together. You shall use hull() with both balls in connection, BUT with each one of them APART. For better understanding please see the picture below.
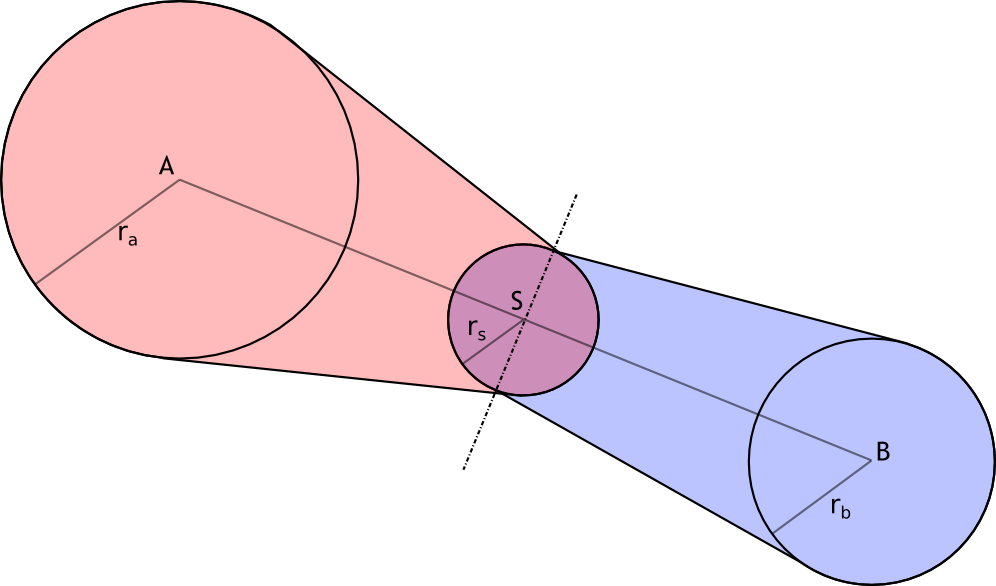
A and B are centers of two balls in linkage. Point S lays in middle of abscissa AB. Radius of middle ball is half of arithmetical average of edge balls. Both of the edge balls will be hulled by the middle separately.
Linkages can go across each others. All amino-acids has at least one linkage. It’s forbidden to manipulate with $fn and $fs variables!
This is example of module which process arguments from vectors:
module echoprotein(aminos=[],bindings=[]) {
for (binding = bindings) {
echo("Koule A:");
echo("x", aminos[binding[0]][0]);
echo("y", aminos[binding[0]][1]);
echo("z", aminos[binding[0]][2]);
echo("r", aminos[binding[0]][3]);
echo("Koule B:");
echo("x", aminos[binding[1]][0]);
echo("y", aminos[binding[1]][1]);
echo("z", aminos[binding[1]][2]);
echo("r", aminos[binding[1]][3]);
}
}Example of income arguments:
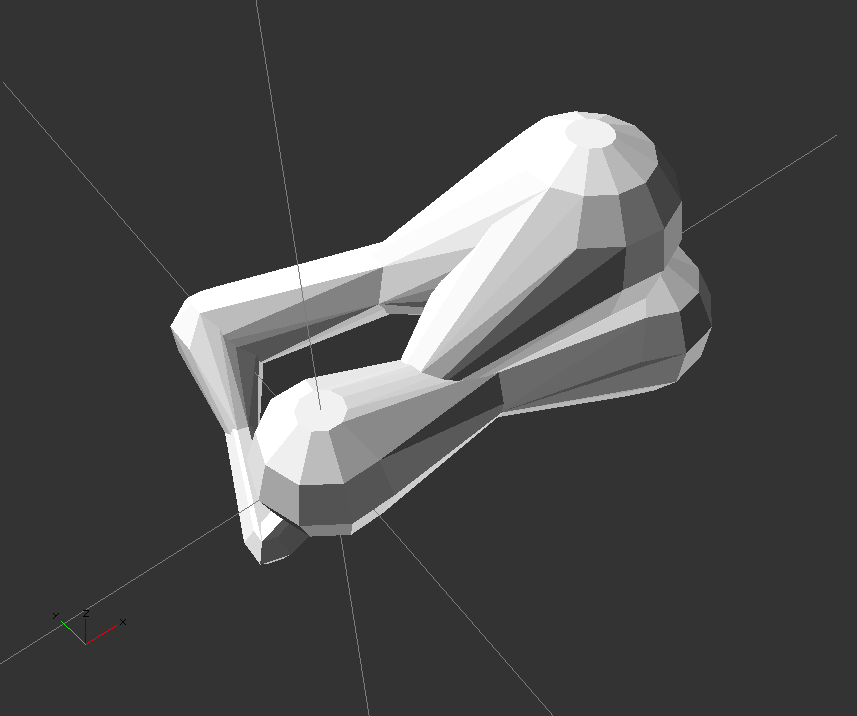
protein([
[0,0,0,3],
[12,0,5,4],
[14,-2,0,3.5],
[-3,0,-3,1],
[0,8,0,2],
],
[
[0,2],
[1,3],
[1,4],
[3,4],
]
);

Your Proteins
You have been assigned to 3 different automatically generated proteins. You have them as raw data arguments for module protein(), you have them also as .STL files. Most probably you will not be able to render the model on your computer within some time interval which will not be too long. You shall use subset of these data to test your solution.
Your proteins are accessible on these addresses:
You need to replace XXX with digits from this list:
xxx 045 001 002 ...
Pre-processing
Choose which ever protein out of the 3 that has been given to you and prepare it for print. You can do what ever you want with this model (rotate, cut, fix, add support), but it is neccessary to keep dimensions of protein. Output of this section is one or more STL files, which are ready for slicing, and very brief description of the things which you did with the original model and why. (This brief description is not to be written but you need to explain all the steps to the person which will check your model).
Slicing
Slice it by which ever program you prefer, you can also develop you own slicer if you like. For Slic3r use slic3r-config-bundle.ini.zip (hot-end 3dráty, filament 1.75 ABS), for KISSlicer are here, you need to change some parameters in configs to get the print right, do not use them as they are! After you slice your model, you need to export used config, you also need to keep sliced model in .GCODE. Fit all your objects on one plate and subbmit to us one GCODE file!
Print and post-proccessing
You can find the dates and times for printing in KOS. You will be printing your gcode in our lab. You need to post-proccess printed object afterward (remove supports, glue it together etc.). The print should look like the given model.
In case total failure it is possible to retry with new .GCODE, but only once. In case of failure which is not from you doing the try is not considered.
Submission, evaluation and terms
Upload .ZIP archive to EDUX, link will point to this location from your namespace. Every additional text please put to your namespace.
What do you submit?
- scad file containing modul
protein - scad file with call to your modul with arguments
- STL file with your chosen protein
- STL file which you will print
- GCODES
- Slicer config you used
- Any necessary files we need to evaluate you work
Last possible date to submit you work is midnight the day before you go to print the work.
Points according to this:
| OpenSCAD module | 10 | |
|---|---|---|
| Modul works accordingly the task | 7 | mandatory |
| Source code is well written and accordingly commented | 3 | |
| Preparation for printing | 10 | |
| Well prepared STLs | 5 | mandatory |
| Mesh is valid in all STL files | 5 | |
| Slicing | 10 | |
| Supports (are not necessary (5 p.), justified use* (2.5 p.), unjustified use (0 p.)) | 5 | |
| Well chosen parameters for printing (perimeters, infill, height of layer) | 5 | |
| 20 | ||
| The printed model is accordingly by task, the object is post-proccessed | 5 | mandatory |
| The printed model does not have any defects caused by wrong preparation it self for printing | 7.5 | |
| The printed model does not have any defects caused by wrong usage of printer (preparation of heat bed, bad temperature) | 7.5 |
- We lower your points only fot automatically generated support (by slicers).
Evaluation is divided to 4 parts. mandatory means that if you fail in this one you will not receive any points from the section it is in!